Welcome to AI Drug Lab!
Artificial intelligence (AI) has been widely applied in drug discovery. Various machine learning and deep learning models are proved to be capable to provide insights of drug candidates. Encouraged by this success, AI Drug Lab is designed as an integrated platform to conduct AI assisted drug design. AI Drug Lab develops and hosts multiple models online and aims to facilitate the research in drug development and optimization.
Functionalities
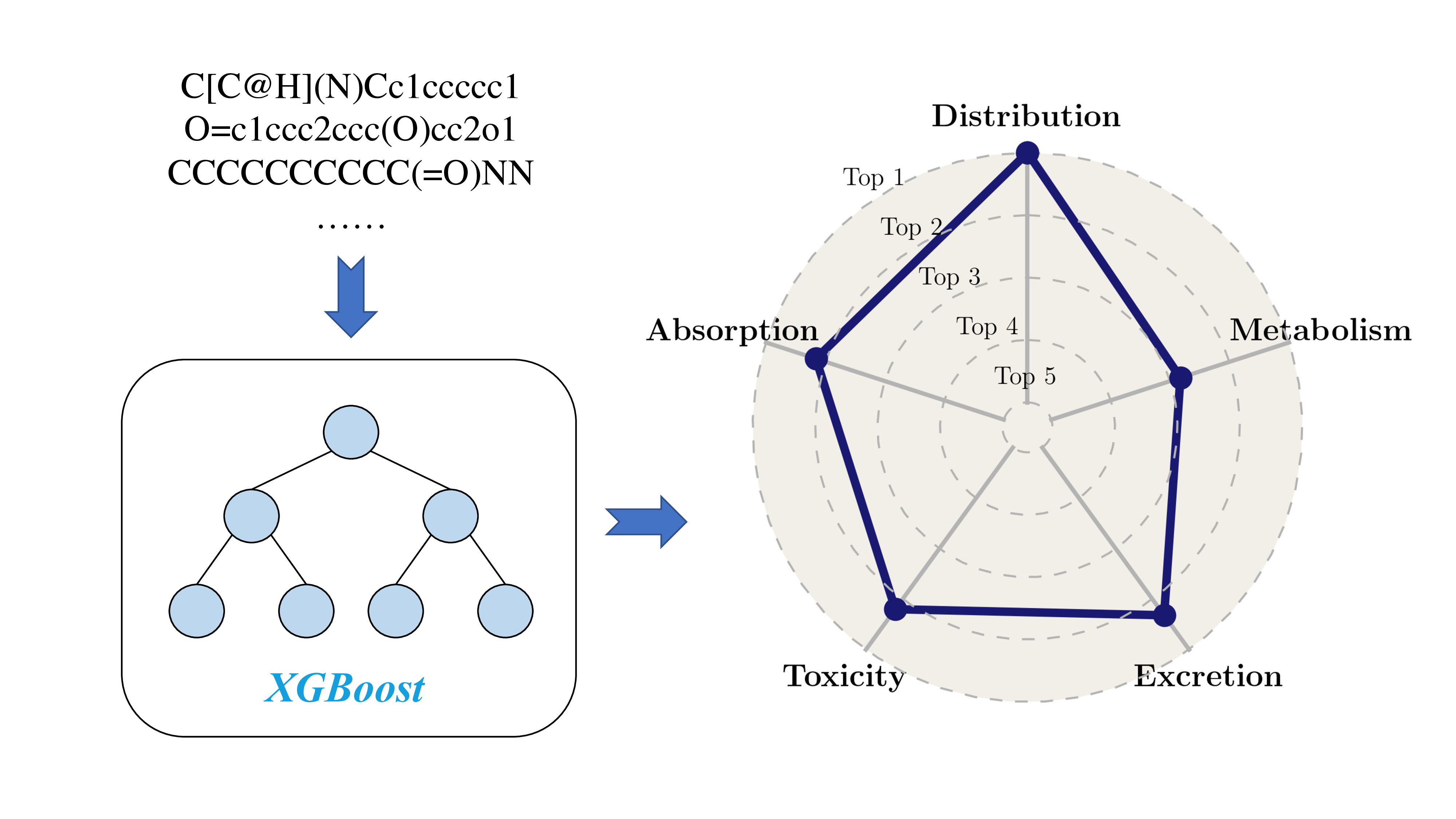
ADMET Prediction
An ensemble of features, including fingerprints and descriptors, and a tree-based machine learning model, extreme gradient boosting, are used for accurate ADMET prediction.
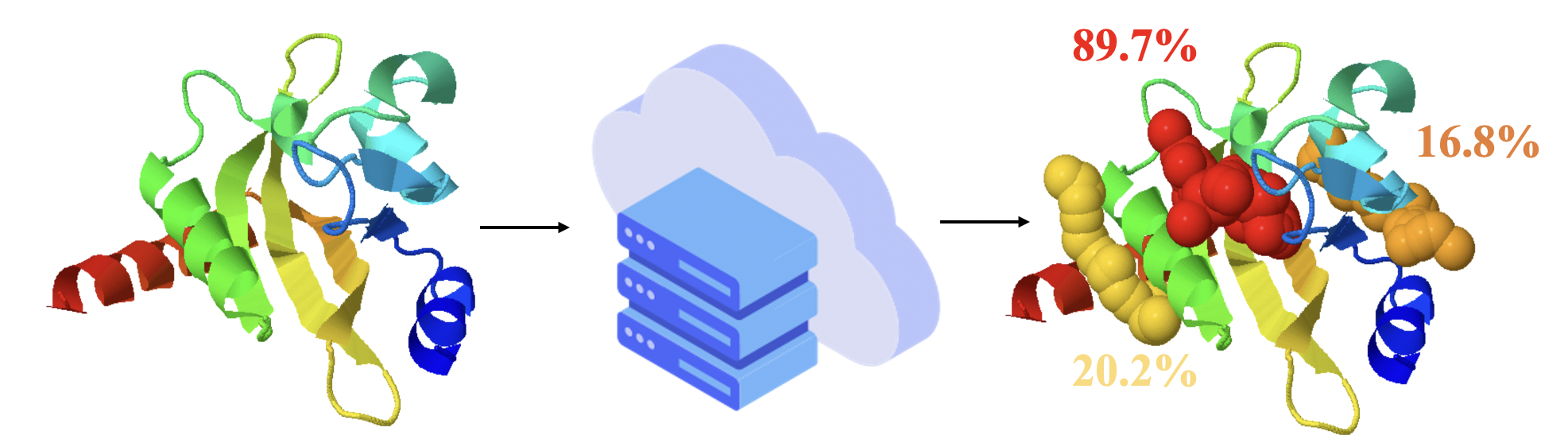
Protein Allosteric Sites Prediction
Protein Allosteric Sites Server (PASSer) is designed for allosteric site prediction. An ensemble learning, consisting of extreme gradient boosting and graph convolutional neural network, is used to learn the physical properties and topology information.
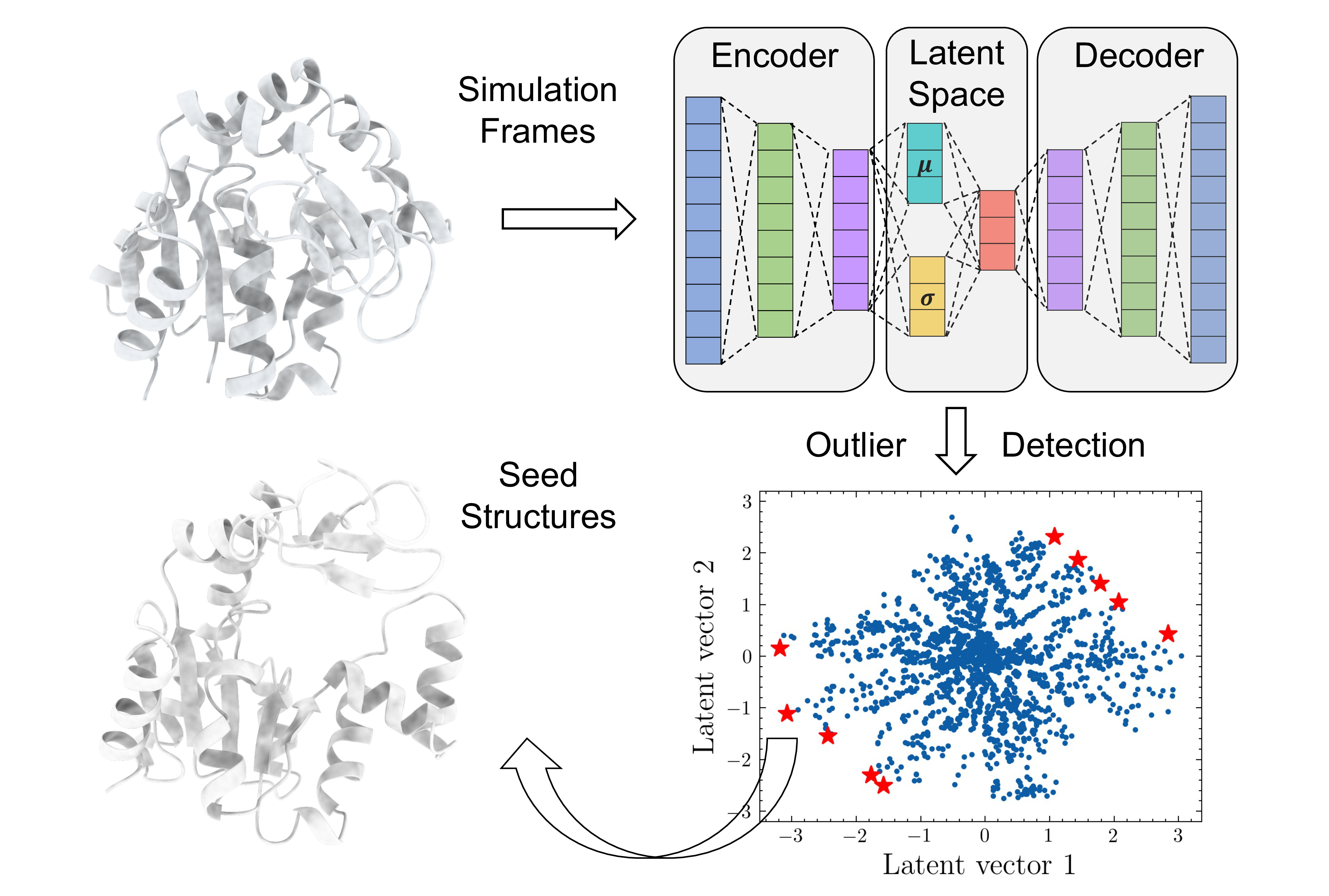
Latent Space Assisted Adaptive Sampling for Protein Trajectories
Latent Space Assisted Adaptive Sampling for Protein Trajectories (LAST) is an adaptive sampling method to accelerate the exploration of protein conformational space This method comprises cycles of (i) variational autoencoders training, (ii) seed structure selection on the latent space and (iii) conformational sampling through additional MD simulations.
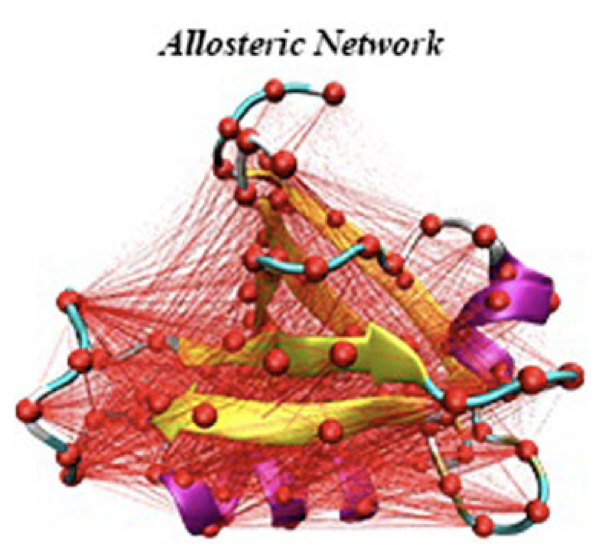
Relative Entropy-based Dynamical Allosteric Network
Relative Entropy-based Dynamical Allosteric Network Model (REDAN) can evaluate protein allostery through relative entropy at three mutually dependent structural levels: allosteric residues, allosteric pathways, and allosteric communities.